Exponential progress in sequencing and the basis of precision medicine
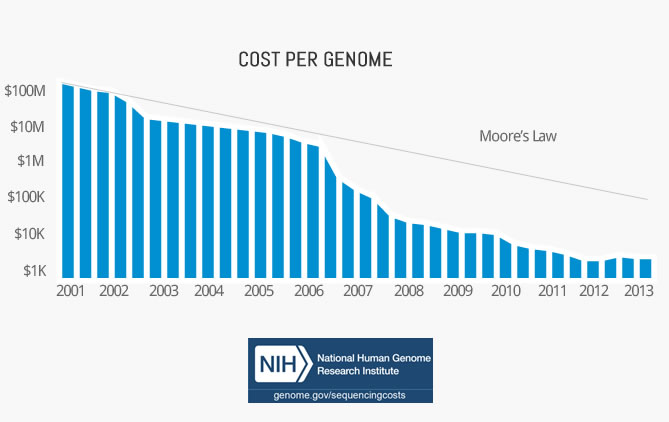
Cost per genome has declined exponentially
Your genome contains the information that forms you. It consists of 3.2 billion base pairs and around 22.000 genes. Knowing this code of life and understanding what it does is crucial for applications ranging from medicine to agriculture. Only when we know what the genome contains, can we use tools such as CRISPR to edit it.
This post is part 2 of my series about exponential technologies in healthcare. Read part 1 here.
The first complete human genome was sequenced by Craig Venter in 2001. The whole project cost $3b, which translates to around $1 per base pair. In total, it took over a decade to complete.
Since then the cost per genome has fallen faster than Moore’s Law. In fact, it outpaced it by almost 3x.
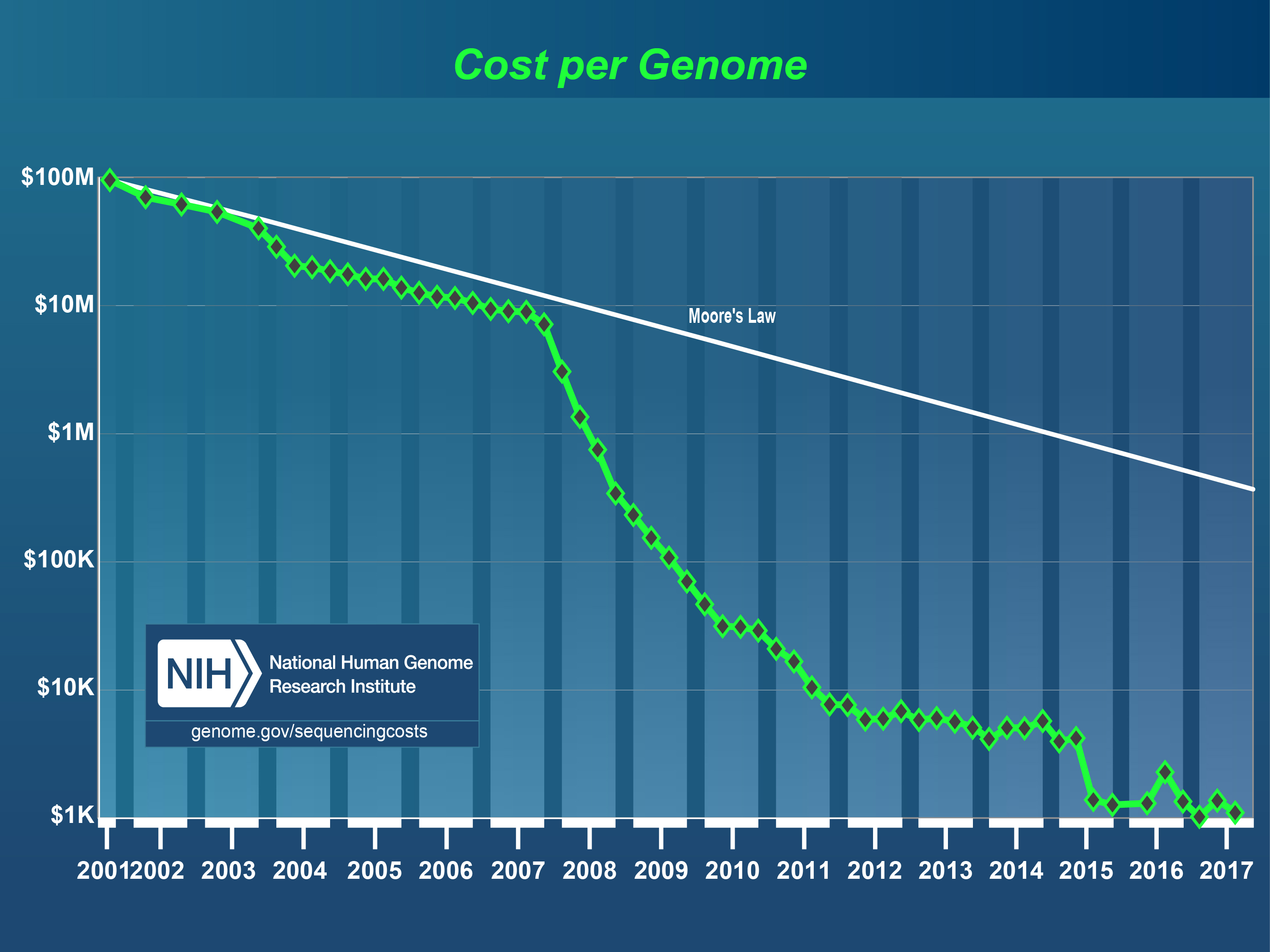
Today, the cost of goods is only a couple hundred dollars, or 0.0000001 USD per base pair. And in a couple of years, it might be below 100 USD in total. That would be cheaper than many routine medical tests today.
Crucially, the time to sequence a genome has fallen significantly as well. State of the art sequencing machines can complete a whole genome in 25 minutes: The time it’ll take you to bake a pizza.
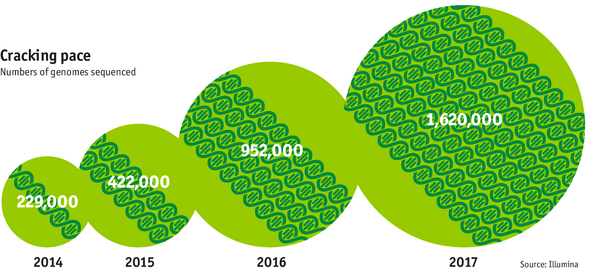
The exponentially falling cost of sequencing has enabled the exponential generation of genomic information. In the end of 2014, around 229.000 genomes have been sequenced in total - up from around 4 in 2008. The generation of genomic information is expected to double every year reaching over 1.6M in 2017.
Plenty productive use cases
This incredible development enables some very exciting and productive new things.
Data Mining
Once we have information on millions of genomes available, we can combine this data with “phenotypic” data from health records, MRIs or tracking devices.
We can also sequence intermediary genomic information such as RNA and add information on the other “OMs” such as the proteome or microbiome.
Using machine learning on these datasets that contain input (gene) and output (health) will allow us to detect correlations and relationships that were unknown before.
We can use all this information to guide research or to predict health and detect diseases.
Segment-of-one care
Today care relies on averages that come from clinical trials. However, most people are not the average in some form. Being able to understand the genome of one particular patient might allow us to target the care exclusively to this one patient. This is the promise of precision medicine.
Molecular drivers of disease
Not so long ago, cancer was classified by organs. There war lung cancer, kidney cancer or breast cancer. Before molecular mechanisms were known, cancer could only be observed and accounted to the organ it befell.
With the advance in sequencing technology, we’ve gained a much more fine-grained picture of cancer on the molecular level. Each cancer now has an individual fingerprint.
It’s DNA and mutations affect the therapeutic choice much more than the organ that it hosts. The approach to choosing a therapy has fundamentally shifted!
Using this kind of information for actually driving decisions is only made possible through the advancements in algorithms and data processing.
Structural understanding
NGS is not only used to sequence the genome. It is also used on all other “OMs”, such as the microbiomed. One use case that I find particularly important is the transcription of the RNA.
In the cell different kinds of RNA directly reflect the gene expression. The RNA transcript represents the second layer of information.
In parallel to the genetic code for protein synthesis, a second layer of information is embedded in all RNA transcripts in the form of RNA structure. RNA structure influences practically every step in the gene expression program. However, the nature of most RNA structures or effects of sequence variation on structure are not known.
Through NGS it is now possible to determine the RNA structure directly across all RNA in the cell. This has revolutionised the whole field. Now scientists don’t need to model the structure by calculating e.g. the binding properties, they can simply measure it. This dramatically improves our understanding of what is going on in the cell.
Noninvasive Prenatal Testing
With NGS we can now analyse the blood of the mother in order to reliably determine genetic problems with the baby. Some of the baby’s DNA is circulating in the mother’s blood. This DNA is sequenced and can reveal problems in the DNA, such as Down Syndrom. The benefits over previous methods are that it can be done earlier and doesn’t put the developing baby at risk.
NGS: Next generation sequencing
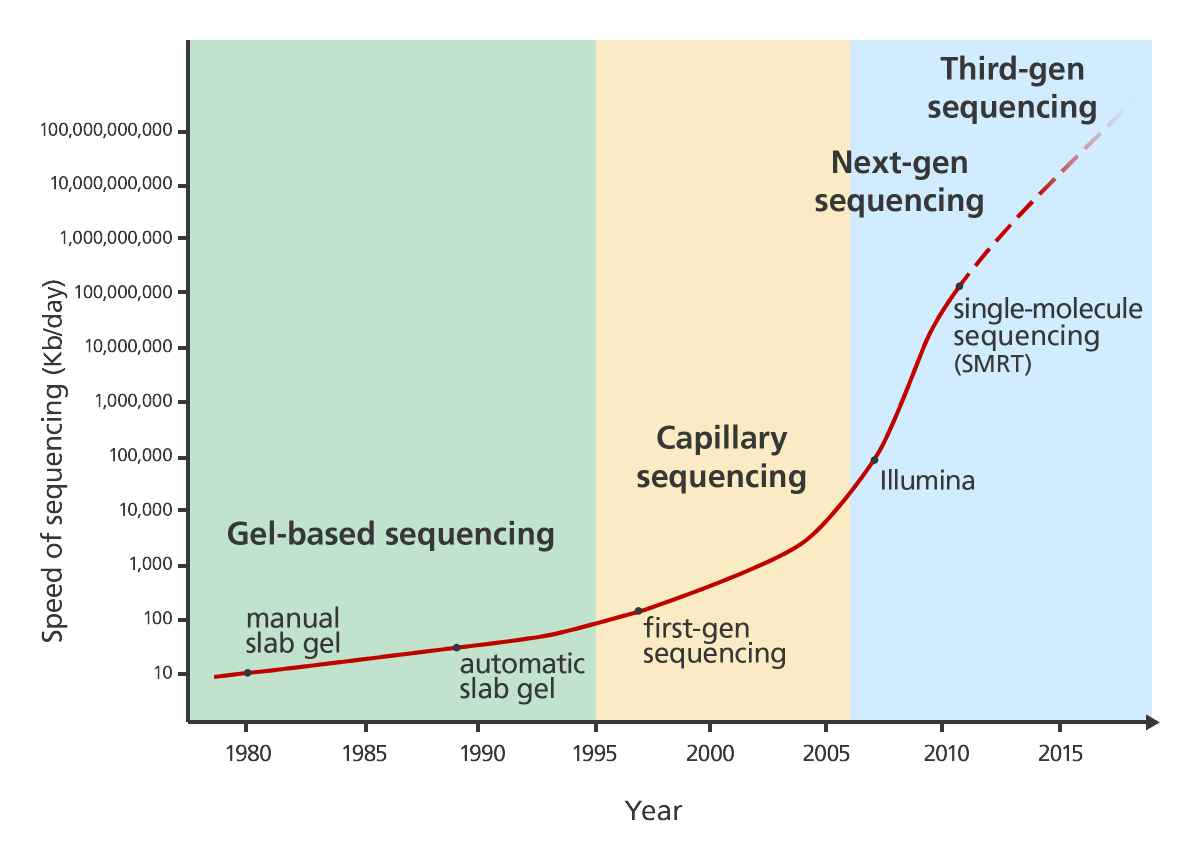
Sequencing technology has dramatically improved and changed over the last decades. It was first developed in the 1970s by Frederick Sanger. This technology was applied to sequence the first human genome. There’s a really nice and explanation of first generation sequencing on Luke Jostins blog.
Several different approaches to next generation sequencing exist. Their advantage is that they work much faster than the Sanger method. One way this speed improvement is achieved is by parallelisation. You can read up on several approaches in another nice blog post by Luke Jostens.
The latest step in NGS is single molecule sequencing. It works by observing the DNA polymerase while it works on the DNA strand nucleotide by nucleotide.
DNA arrays
DNA arrays were the intermediary step in genome sequencing until whole genome sequencing became viable. Companies such as 23andme use this kind of technology to provide genomic analysis.
Instead of sequencing all 3b base pairs, DNA arrays analyse genes in millions of pre-defined places. Chopped up DNA washes over a chip that contains thousands of pre-defined DNA sequences called probes. It then detects if a DNA fragment has bound to any of the probes.
This limited data collection makes this technology much more affordable. For instance 23andme claims to analyse hundreds of thousands of places in the DNA.